PI3K /AKT Signalling pathway and Human Diseases
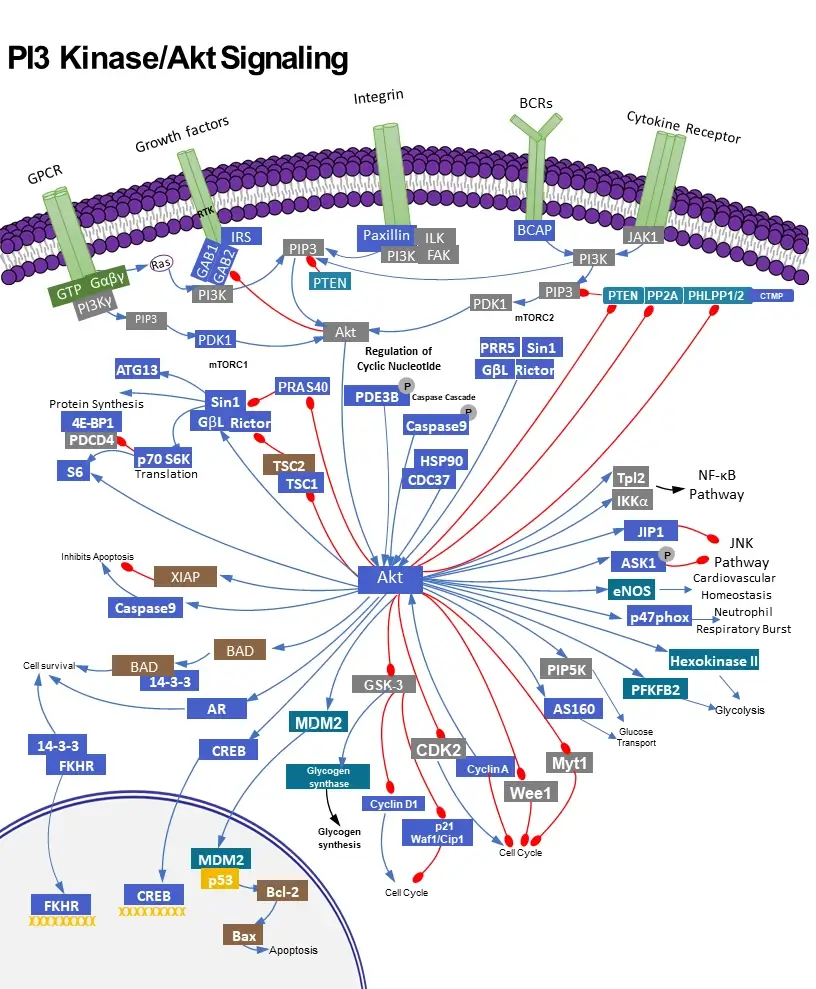

Key functions of PI3K /AKT signal pathway
The PI3K (phosphatidylinositol-3-kinase or phosphatidylinositol-4,5-bisphosphate 3-kinase) signal transduction pathway is utilized ubiquitous among cells and regulates a variety of functions in the body. As such, dysfunction of the PI3K signaling pathway has been implicated in several health problems, such as insulin resistance, diabetes, inflammatory disease, and cancer. Class I PI3K activity leads to activation of the proto-oncogene AKT (Protein Kinase B, PKB), which regulates aspects of the cell cycle including growth, proliferation, and differentiation. Thus research into PI3K-AKT transduction pathways holds great potentials for better understanding and treatments of human cancers.
Key molecules involved in PI3K /AKT signal pathway
Class I PI3Ks are heterodimeric proteins composed of one of four catalytic p110 isoforms (p110α, p110β, p110γ, and p110δ) and one of five p85 regulatory isoforms (p85α, p55α, p50α, p85β, and p55γ). The p110α and p110β subunits are expressed throughout the body while p110γ and p110δ are expressed predominantly in leucocytes. The variety of activation mechanisms of PI3K are complex, being both subunit- and cellular context-dependent. In brief, PI3K is activated by growth factor or hormone binding to RTK (receptor tyrosine kinases) or by Ras-related GTPases and heterotrimeric G proteins. Factors that activate PI3K include, but insulin, epidermal growth factor, platelet-derived growth factor, and cytokines. Different p85 isoforms can modulate the localization and activity of the PI3K heterodimer. The p85 subunit binds and blocks the functional domains of the p110 subunit until activation/phosphorylation causes a conformational change in p85 that uncovers the catalytic domains allowing kinase activity. With such a broad array of activating molecules, mechanisms, and subunits, it is not surprising the PI3K coordinates a variety of downstream effects.
AKT is the most ubiquitous downstream effector of PI3K. Once activated, PI3K phosphorylates the 3’-hydroxyl group of the inositol ring of the plasma membrane phospholipid PIP2 (phosphatidylinositol-4,5-bisphosphate), converting it to PIP3 (phosphatidylinositol-3,4,5-triphosphate) which in turn activates effector proteins, like AKT. AKT is a subfamily of serine/threonine kinases that includes AKT1, AKT2, and AKT3. AKT activation occurs by colocalization of AKT and its activating kinase PDK-1 to the plasma membrane. Activated AKT has numerous targets involved in cell proliferation, differentiation, metabolism, and trafficking [1]. Among the downstream effects of AKT is increased glucose uptake via translocation of glucose transporters (GLUT1 and GLUT4) to the plasma membrane and increased glycogen synthesis via inhibition of glycogen synthase kinase 3 (GSK3). AKT also increases protein synthesis through activation of mTORC1 (mammalian target of rapamycin complex 1) that facilitates translation at ribosomes. An additional important function of AKT is marking the FOXO (Forkhead box O) transcription factor for degradation, leading to increased cell survival and proliferation. The proteins PTEN, PP2A, and PHLPP down-regulate AKT activation. PTEN in particular, through down-regulation of AKT as well as via other pathways, serves as an important tumor suppressor.
PI3K /AKT signaling pathway in human disease
Aberrant activation of the PI3Ksignalling pathway occurs in most human cancers. The potential for aberrant over-activity of PI3K to transform healthy cells to cancerous cells has been known for decades (e.g.: [2], [3]). In 2004 Samuels et al. [4] found activating mutations of the p110α subunit gene, PIK3CA, in human cancer cells, and mutations of this gene are associated with a wide array of cancers (see: www.tumorportal.org). The p110α subunit has been associated with a variety of cancers while mutations of other p110 genes are not commonly associated with human cancers. However, p85 subunit truncations which reduce the regulatory contacts with p110α can lead to transformation following p110α activation [5]. Further, loss-of-function mutations of the AKT inhibiting PTENgene are frequently present in human cancers. Mutations of PIK3CA and PTEN are two of the most frequent mutations among all human cancers [6].
PI3K activation by insulin along with the effects of this pathway on glucose uptake and metabolism makes it a strong candidate for involvement in the development of insulin resistance and diabetes. Data suggest that genetic variation within the PI3K-AKT-mTOR pathway is associated with the development of type 2 diabetes [7]. PI3K and AKT increase mTORC1 activity, stabilizing the protein GRB10, which then binds to and down-regulates the insulin receptor. Variants of the GRB10 gene have also been linked to increased risk of type 2 diabetes [8]. Additionally, liver-specific p110α knockout in mice leads produces insulin resistance that can be rescued by overexpression of p110β [9]. We are far from a full understanding of the role(s) of PI3K signal transduction in diabetes and insulin sensitivity, but it is clear that it complex and likely involves both AKT-dependent and AKT-independent pathways. Furthermore, the sort of “tug-of-war” of PI3K signaling up-regulation involved in health conditions like tumorogenesis and down-regulation in glucose regulation/metabolism, both of which typically involve p110α, complicates the task of developing treatments that target this pathway.
The PI3K signaling pathway’s involvement in a wide array of human health dysfunctions makes proteins in this pathway exciting potential targets for treating a variety of disease states. However, the prospect of developing such treatments is more difficult than initially realized. A variety of PI3K proteins with overlapping functions exist within cells and are activated by a wide array of receptor proteins. This complicates both identifying the exact source of dysfunction in a given patient and attempts to target one protein without compensatory regulation of related proteins. Additionally, there are important health problems caused by both under- and over-activity of PI3K signaling so that a targeted therapy that pushes activity too far in one direction can produce other health problems. Furthermore, PI3K pathway inhibitors applied in vitro to cancer cell lines often induces adaptive resistance to the inhibitors within days [10].
In spite of these difficulties, the PI3K pathway holds hope for treatment of cancers and other human diseases. Personalized medicine is rapidly advancing and the ability to target specific mutational variants holds great potential. Research continues to elucidate the many complexities of PI3K signaling pathways, advancing us towards new therapies and, importantly, uncovering unrealized roles in human health.
 Loading ....
Loading ....
