Optimizing Sequencing Costs in Metagenomics

How to reduce costs of pathogen detection in with PaRTI-Seq™
Pathogen detection techniques based on culture PCR, or serology are still regarded as the gold standards for pathogen detection. However, these techniques present significant limitations due to their turnover time, low positivity rates and low sensitivity. Next-generation sequencing (NGS) techniques can address many of these shortcomings with the high-throughput approach of nucleic acid sequencing, in which millions to billions of short fragments of DNA (reads) or RNA (transcripts) are massively sequenced in parallel (Behjati and Tarpey, 2013). With the help of bioinformatics, these fragments can then be assembled and mapped against a reference database for accurate and rapid detection of pathogens. NGS-based approaches have the potential to significantly improve throughput capabilities of clinical laboratories, while also enhancing detection sensitivity and resolution.
NGS-based technologies are becoming increasingly affordable per megabase of DNA, but sequencing costs still represent a significant portion (up to 80%) of the total costs associated with their use.
While the cost of sequencing and turn-around times (TAT) associated with NGS have been consistently decreasing (Wetterstrand, 2021), host DNA interference still presents a major limitation that withholds NGS from mass adoption in clinical settings today. A major driver for the reduction of cost and TAT of the sequencing run is an effective host depletion method. Micronbrane's DevinTM Filter (BHE17500003) and PaRTI-SeqTM (Pathogen Real-Time Identification by Sequencing) products provide healthcare providers with a novel, comprehensive solution that enables NGS pathogen detection for clinical diagnostics of infectious diseases to be more efficient and affordable (Figure 1).
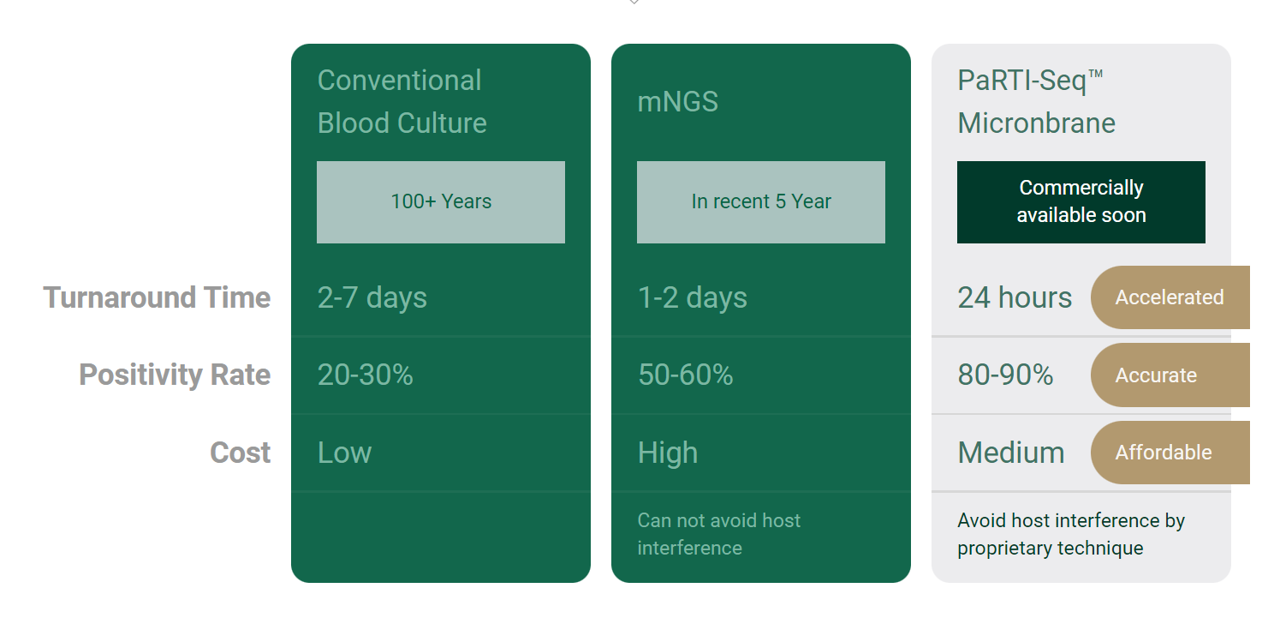
Figure 1. Different methods for the detection of pathogens
PaRTI-SeqTM is a novel next sequencing assay built with DevinTM Filter and Micronbrane’s proprietary database and data analysis pipeline (Figure 2). It offers an accelerated, affordable, and accurate solution for pathogen identification in clinical samples. It can save up to 75% of sequencing costs and reduce more than half of overall costs for such tests in clinical settings while providing accurate results within 24 hours (Figure 3).
.png)
Figure 2. PaRIT-SeqTM workflow
DevinTM Filter (BHE17500003), DNA Enrichment Kit (BHE17500001), NGS Library Prep Kit (BHE17500004)
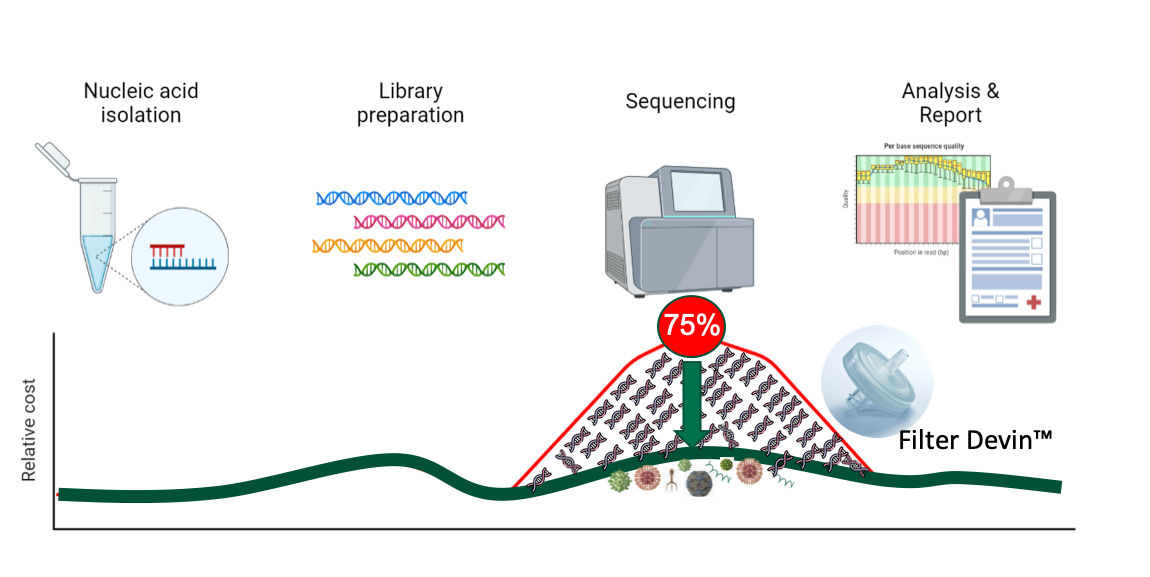
Figure 3. PaRTI-Seq™ reduces 75% of sequencing costs
Download the white paper of PaRTI-Seq™
Watch how it works >>
Watch online animation protocol >>
See all the products by Micronbrane Medical >>
Learn more about our Partner
Micronbrane Medical is a biotechnology company based in Taiwan. They provide cost-effective, rapid, and accurate pathogen detection solutions for human bloodstream infections with their patented host depletion technology and proprietary analytical methods.
 Loading ....
Loading ....
